Unmasking Quinolone Resistance in Environmental Bacteria: How Environmental Reservoirs Are Fueling a Global Antimicrobial Crisis. Discover the Mechanisms, Impacts, and Urgent Solutions Needed to Combat This Growing Menace. (2025)
- Introduction: The Rise of Quinolone Resistance in Environmental Bacteria
- Mechanisms of Quinolone Resistance: Genetic and Biochemical Insights
- Environmental Reservoirs: Sources and Hotspots of Resistance Genes
- Transmission Pathways: From Environment to Human and Animal Health
- Detection and Surveillance: Current Technologies and Methodologies
- Case Studies: Global Incidents and Regional Trends
- Impact on Public Health and Ecosystems
- Regulatory and Policy Responses: International and National Initiatives
- Emerging Technologies and Future Solutions
- Forecasting the Future: Trends, Public Awareness, and the Road Ahead
- Sources & References
Introduction: The Rise of Quinolone Resistance in Environmental Bacteria
Quinolones, a class of broad-spectrum antibiotics, have been widely used in human medicine, veterinary practice, and agriculture since their introduction in the 1960s. Their extensive application has contributed to the emergence and proliferation of quinolone-resistant bacteria, not only in clinical settings but increasingly in diverse environmental compartments. As of 2025, the rise of quinolone resistance in environmental bacteria is recognized as a critical public health concern, with implications for the efficacy of antimicrobial therapies and the spread of resistance genes across ecosystems.
Recent surveillance and research efforts have highlighted the pervasive presence of quinolone-resistant bacteria in water bodies, soils, and sediments, particularly in regions with high antibiotic usage and inadequate wastewater treatment. Environmental monitoring programs coordinated by organizations such as the World Health Organization and the European Medicines Agency have documented increasing detection rates of resistance determinants, including plasmid-mediated quinolone resistance (PMQR) genes, in environmental isolates. These findings underscore the role of environmental reservoirs in the maintenance and dissemination of resistance traits.
Key events in recent years include the identification of novel resistance mechanisms and the mapping of resistance gene flow between environmental, animal, and human microbiomes. For example, studies supported by the Centers for Disease Control and Prevention have demonstrated that environmental bacteria can serve as a source of resistance genes that may be transferred to clinically relevant pathogens, complicating infection control and treatment strategies. The detection of high levels of quinolone residues in effluents from pharmaceutical manufacturing and agricultural runoff further exacerbates selective pressure, promoting the evolution and persistence of resistant strains.
Looking ahead to the next few years, the outlook for quinolone resistance in environmental bacteria remains challenging. Global health authorities, including the World Health Organization, are calling for enhanced surveillance, stricter regulation of antibiotic use, and improved wastewater management to mitigate the spread of resistance. Advances in molecular diagnostics and metagenomic sequencing are expected to provide deeper insights into resistance dynamics and facilitate the development of targeted interventions. However, without coordinated international action and sustained investment in antimicrobial stewardship, the environmental dimension of quinolone resistance is likely to persist as a significant threat to public health and ecosystem integrity.
Mechanisms of Quinolone Resistance: Genetic and Biochemical Insights
Quinolone resistance in environmental bacteria has become an increasingly pressing concern, particularly as surveillance in 2025 reveals a growing prevalence of resistance determinants outside clinical settings. The mechanisms underlying this resistance are multifaceted, involving both genetic and biochemical adaptations that enable bacteria to survive quinolone exposure. Quinolones, which target bacterial DNA gyrase and topoisomerase IV, are rendered less effective through several well-characterized pathways.
Genetically, the most prominent mechanism involves mutations in the quinolone resistance-determining regions (QRDRs) of the gyrA and parC genes. These mutations alter the target enzymes, reducing drug binding affinity. Recent environmental isolates, particularly from aquatic and soil ecosystems, have shown a notable increase in QRDR mutations, suggesting ongoing selective pressure from environmental contamination with quinolones and related compounds. In addition to chromosomal mutations, plasmid-mediated quinolone resistance (PMQR) genes, such as qnr, aac(6’)-Ib-cr, and qepA, have been detected with rising frequency in environmental samples. These genes can be horizontally transferred between bacteria, facilitating rapid dissemination of resistance traits across diverse microbial communities.
Biochemically, resistance is further enhanced by the upregulation of efflux pumps, such as those encoded by the acrAB-tolC operon, which actively expel quinolones from the bacterial cell. Environmental bacteria, especially those exposed to sub-inhibitory concentrations of antibiotics in wastewater or agricultural runoff, often exhibit increased efflux activity. Additionally, some bacteria produce protective proteins that shield DNA gyrase from quinolone action, a mechanism associated with certain PMQR genes.
Recent data from global monitoring initiatives, including those coordinated by the World Health Organization and the European Food Safety Authority, indicate that environmental reservoirs of quinolone resistance are expanding. These organizations have highlighted the role of environmental bacteria as both indicators and vectors of resistance genes, with implications for human and animal health. The persistence and spread of resistance determinants in the environment are expected to continue over the next few years, driven by ongoing antibiotic use in agriculture, aquaculture, and improper disposal of pharmaceuticals.
Looking ahead, the outlook for quinolone resistance in environmental bacteria remains concerning. The convergence of genetic mobility, biochemical adaptability, and environmental contamination is likely to sustain and even accelerate the emergence of resistant strains. Enhanced surveillance, stricter regulation of antibiotic use, and improved waste management practices are being advocated by international bodies to mitigate this trend. Continued research into the molecular mechanisms of resistance will be crucial for developing novel strategies to curb the environmental spread of quinolone resistance.
Environmental Reservoirs: Sources and Hotspots of Resistance Genes
Quinolone resistance in environmental bacteria has emerged as a critical concern in 2025, reflecting the broader challenge of antimicrobial resistance (AMR) in non-clinical settings. Environmental reservoirs—such as surface waters, soils, sediments, and wastewater—serve as both sources and hotspots for the proliferation and dissemination of quinolone resistance genes (qnr, aac(6’)-Ib-cr, qepA, and others). These genes are often associated with mobile genetic elements, facilitating horizontal gene transfer among diverse bacterial populations.
Recent surveillance data indicate that environmental compartments, particularly those impacted by anthropogenic activities, harbor elevated levels of quinolone-resistant bacteria. Wastewater treatment plants (WWTPs) are recognized as major hotspots, as they receive inputs from hospitals, pharmaceutical manufacturing, and urban runoff. Studies in 2024 and early 2025 have shown that even advanced treatment processes do not fully eliminate resistant bacteria or resistance genes, allowing their release into receiving water bodies. Agricultural soils irrigated with reclaimed water or amended with manure from treated animals also represent significant reservoirs, with quinolone residues and resistance genes persisting and spreading through microbial communities.
The World Health Organization (WHO) and the European Medicines Agency (EMA) have highlighted the environmental dimension of AMR, urging for integrated surveillance and mitigation strategies. The EMA has specifically addressed the environmental risk assessment of veterinary medicinal products, including quinolones, emphasizing the need for stricter controls on environmental emissions. The United States Environmental Protection Agency (EPA) is also advancing research on the fate and transport of antibiotics and resistance genes in aquatic systems, supporting the development of new monitoring frameworks.
In 2025, metagenomic analyses and high-throughput sequencing are increasingly used to map the diversity and abundance of quinolone resistance genes in environmental samples. These approaches have revealed complex networks of gene exchange between environmental, commensal, and pathogenic bacteria, underscoring the interconnectedness of environmental and clinical AMR. The persistence of quinolone residues in the environment, often at sub-inhibitory concentrations, continues to select for resistant strains, raising concerns about the long-term efficacy of this antibiotic class.
Looking ahead, the outlook for controlling quinolone resistance in environmental bacteria depends on coordinated global action. Enhanced regulatory oversight, improved wastewater treatment technologies, and the reduction of unnecessary quinolone use in agriculture and human medicine are key priorities. International organizations, including the World Health Organization, are expected to expand their One Health initiatives, integrating environmental surveillance into broader AMR containment strategies over the next few years.
Transmission Pathways: From Environment to Human and Animal Health
The transmission of quinolone resistance from environmental bacteria to human and animal populations is a growing concern in 2025, driven by the widespread use of quinolone antibiotics in healthcare, agriculture, and aquaculture. Environmental reservoirs—such as surface waters, soils, and wastewater—act as critical hubs for the persistence and dissemination of quinolone-resistant bacteria and resistance genes. These pathways facilitate the movement of resistance determinants across ecological boundaries, ultimately impacting public and animal health.
Recent surveillance data indicate that environmental bacteria, particularly those in aquatic environments, frequently harbor plasmid-mediated quinolone resistance (PMQR) genes, such as qnr, aac(6’)-Ib-cr, and qepA. These genes can be horizontally transferred to clinically relevant pathogens via mobile genetic elements, including plasmids and integrons. The World Health Organization (WHO) has highlighted the role of environmental contamination in the global spread of antimicrobial resistance (AMR), emphasizing the need for integrated monitoring across sectors.
Transmission pathways are multifaceted. Wastewater treatment plants (WWTPs) are recognized as hotspots for the accumulation and release of quinolone-resistant bacteria into natural water bodies. Studies in 2024 and early 2025 have shown that even advanced treatment processes may not fully eliminate resistant bacteria or resistance genes, allowing their entry into rivers and lakes. These contaminated waters can then be used for irrigation, recreation, or as drinking water sources, creating direct and indirect exposure routes for humans and animals.
Agricultural practices further amplify the problem. The use of manure and biosolids as fertilizers introduces quinolone residues and resistant bacteria into soils, where resistance genes can persist and be taken up by soil microbiota. Crops irrigated with contaminated water or fertilized with such materials may serve as additional vectors for resistance transmission. The Food and Agriculture Organization of the United Nations (FAO) has called for stricter regulations on antibiotic use in agriculture and improved waste management to curb environmental AMR dissemination.
Wildlife and companion animals also play a role in the environmental-human interface. Animals exposed to contaminated environments can acquire and spread quinolone-resistant bacteria, acting as reservoirs and vectors. The World Organisation for Animal Health (WOAH, formerly OIE) is actively monitoring AMR in animal populations and promoting a One Health approach to address these interconnected risks.
Looking ahead, the outlook for 2025 and beyond involves strengthening surveillance systems, advancing wastewater treatment technologies, and implementing coordinated policies across human, animal, and environmental health sectors. International organizations are expected to intensify efforts to map transmission pathways and develop targeted interventions, recognizing that environmental reservoirs are pivotal in the ongoing challenge of quinolone resistance.
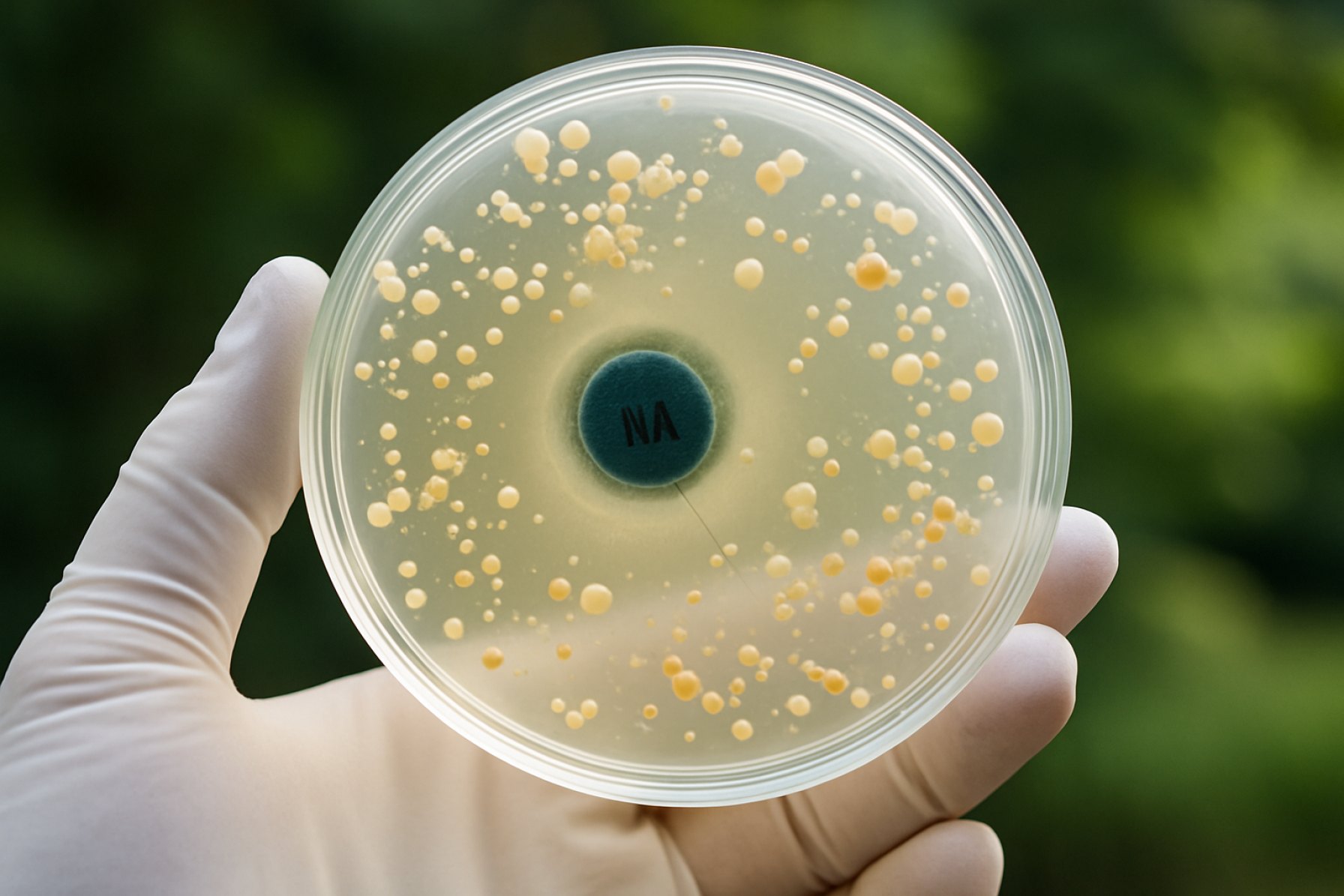
Detection and Surveillance: Current Technologies and Methodologies
The detection and surveillance of quinolone resistance in environmental bacteria have become increasingly sophisticated, reflecting the urgent need to monitor antimicrobial resistance (AMR) beyond clinical settings. As of 2025, a combination of molecular, culture-based, and metagenomic approaches are employed to track the prevalence and dissemination of quinolone resistance genes (qnr, aac(6’)-Ib-cr, qepA, etc.) in diverse environmental matrices such as water, soil, and wastewater.
Polymerase chain reaction (PCR) and quantitative PCR (qPCR) remain the cornerstone for the rapid detection of known quinolone resistance determinants. These methods allow for high sensitivity and specificity, enabling the quantification of resistance genes in complex samples. Recent advances include multiplex PCR assays that can simultaneously detect multiple resistance genes, streamlining surveillance efforts. Whole genome sequencing (WGS) and metagenomic sequencing have gained traction, providing comprehensive insights into the resistome of environmental samples and uncovering novel resistance mechanisms. These high-throughput sequencing technologies are increasingly accessible due to declining costs and improved bioinformatics pipelines, facilitating large-scale surveillance projects.
Culture-based methods, while more labor-intensive, are still essential for isolating viable resistant bacteria and performing phenotypic susceptibility testing. These methods are often used in conjunction with molecular techniques to validate findings and assess the clinical relevance of detected resistance genes. Selective media containing quinolones are commonly used to enrich for resistant strains from environmental samples.
Automated platforms and portable devices are emerging as valuable tools for on-site detection. For example, portable qPCR instruments and isothermal amplification technologies (such as LAMP) are being deployed for rapid field-based surveillance, particularly in resource-limited settings. These innovations are expected to expand in the next few years, improving the timeliness and geographic reach of environmental AMR monitoring.
International organizations such as the World Health Organization and the Centers for Disease Control and Prevention have emphasized the importance of environmental surveillance in their AMR action plans. The European Centre for Disease Prevention and Control is also supporting harmonized surveillance protocols across member states. These agencies are promoting the integration of environmental data into national and global AMR surveillance systems, recognizing the environment as a critical reservoir and transmission route for quinolone resistance.
Looking ahead, the next few years are likely to see further integration of real-time data analytics, machine learning, and geospatial mapping into surveillance platforms. This will enhance the ability to detect emerging resistance hotspots and inform targeted interventions. The continued development of standardized methodologies and international data sharing frameworks will be crucial for effective global surveillance of quinolone resistance in environmental bacteria.
Case Studies: Global Incidents and Regional Trends
The global emergence and spread of quinolone resistance in environmental bacteria have become increasingly evident through a series of case studies and regional surveillance reports. In 2025, several key incidents and trends highlight the complexity and urgency of this issue.
In Asia, particularly in China and India, environmental monitoring has revealed high levels of quinolone-resistant bacteria in surface waters, agricultural soils, and effluents from pharmaceutical manufacturing. Studies have shown that rivers receiving untreated or partially treated wastewater from antibiotic production facilities harbor Escherichia coli and Pseudomonas species with plasmid-mediated quinolone resistance genes (PMQR), such as qnr and aac(6’)-Ib-cr. These findings underscore the role of industrial discharge and inadequate wastewater treatment in amplifying resistance reservoirs in the environment.
In Europe, the European Medicines Agency and the European Food Safety Authority have coordinated surveillance programs that track antimicrobial resistance in environmental samples, including water bodies near livestock farms and urban centers. Recent data indicate a rising prevalence of quinolone-resistant Enterobacteriaceae in river sediments and agricultural runoff, particularly in regions with intensive animal husbandry. The detection of resistance genes in wildlife and migratory birds further suggests environmental dissemination beyond direct human or agricultural sources.
In North America, the Centers for Disease Control and Prevention (CDC) and the United States Environmental Protection Agency (EPA) have reported sporadic but concerning outbreaks of quinolone-resistant bacteria in recreational waters and municipal wastewater. These incidents have prompted local authorities to enhance monitoring and implement stricter guidelines for antibiotic disposal and wastewater management.
Africa and South America are experiencing growing challenges due to limited infrastructure for wastewater treatment and antibiotic stewardship. Surveillance by the World Health Organization (WHO) has documented the spread of quinolone resistance in environmental isolates from rivers and lakes, often linked to informal settlements and unregulated pharmaceutical use.
Looking ahead, the outlook for quinolone resistance in environmental bacteria remains concerning. The continued expansion of urbanization, agricultural intensification, and global trade are expected to facilitate further dissemination of resistance genes. International organizations, including the WHO and the World Organisation for Animal Health (WOAH), are calling for integrated One Health approaches that combine environmental, human, and animal health surveillance to curb the spread of resistance. Enhanced regulatory frameworks, investment in wastewater treatment, and global data sharing are likely to be key priorities in the next few years.
Impact on Public Health and Ecosystems
The proliferation of quinolone resistance in environmental bacteria is an escalating concern for both public health and ecosystem integrity, with significant implications projected for 2025 and the near future. Quinolones, a class of broad-spectrum antibiotics, are widely used in human medicine, veterinary practice, and agriculture. Their extensive application has led to the dissemination of quinolone residues and resistant bacteria into various environmental compartments, including surface waters, soils, and sediments.
Recent surveillance data indicate that environmental reservoirs—such as wastewater treatment plants, agricultural runoff, and natural water bodies—are increasingly recognized as hotspots for the emergence and spread of quinolone-resistant bacteria. These environments facilitate horizontal gene transfer, enabling resistance genes to move between environmental and pathogenic bacteria. The World Health Organization (WHO) has highlighted the environmental dimension of antimicrobial resistance (AMR) as a critical area for intervention, noting that environmental bacteria can serve as a reservoir for resistance genes that may ultimately compromise the efficacy of quinolones in clinical settings.
The public health impact is multifaceted. First, the presence of quinolone-resistant bacteria in the environment increases the risk of human exposure through recreational water use, consumption of contaminated food, and direct contact with animals. This can lead to infections that are more difficult to treat, requiring alternative or more toxic antibiotics. Second, the environmental spread of resistance genes can undermine infection control efforts in healthcare and community settings. The Centers for Disease Control and Prevention (CDC) has underscored the threat posed by resistant pathogens, particularly in vulnerable populations such as immunocompromised individuals.
Ecosystem impacts are also significant. Quinolone residues and resistant bacteria can disrupt microbial communities essential for nutrient cycling, soil fertility, and water quality. The United States Environmental Protection Agency (EPA) and similar agencies globally are increasingly monitoring antibiotic residues and resistance markers in environmental matrices, recognizing their potential to alter ecosystem functions and biodiversity.
Looking ahead, the outlook for 2025 and beyond involves a concerted effort to strengthen environmental surveillance, implement stricter regulations on antibiotic use, and promote the development of advanced wastewater treatment technologies. International collaborations, such as those coordinated by the Food and Agriculture Organization of the United Nations (FAO) and the World Organisation for Animal Health (WOAH), are expected to play a pivotal role in addressing the environmental dimensions of quinolone resistance. Without effective interventions, the continued spread of resistance in environmental bacteria poses a growing threat to both public health and ecosystem sustainability.
Regulatory and Policy Responses: International and National Initiatives
The growing concern over quinolone resistance in environmental bacteria has prompted a range of regulatory and policy responses at both international and national levels, particularly as the global community enters 2025. Quinolones, a class of broad-spectrum antibiotics, are widely used in human medicine, veterinary practice, and agriculture. Their extensive use has contributed to the emergence and dissemination of resistant bacteria in various environmental compartments, including water bodies, soil, and wildlife. This has significant implications for public health, as environmental reservoirs can facilitate the transfer of resistance genes to clinically relevant pathogens.
At the international level, the World Health Organization (WHO) continues to play a central role in coordinating global efforts to combat antimicrobial resistance (AMR), including quinolone resistance. The WHO’s Global Action Plan on AMR, first adopted in 2015, remains a guiding framework for member states, emphasizing the need for surveillance, stewardship, and research. In 2024 and into 2025, the WHO has intensified its focus on environmental dimensions of AMR, urging countries to monitor antibiotic residues and resistant bacteria in the environment and to develop national action plans that address environmental sources of resistance.
The Food and Agriculture Organization of the United Nations (FAO) and the World Organisation for Animal Health (WOAH, formerly OIE) are also key actors, particularly regarding the use of quinolones in food-producing animals and aquaculture. These organizations have issued updated guidelines and recommendations for prudent antibiotic use, and in 2025, they are expected to further strengthen their monitoring and reporting requirements for member countries. The FAO, for example, is expanding its AMR monitoring programs to include environmental sampling in agricultural settings.
At the national level, regulatory responses vary but are increasingly converging toward stricter controls. The European Medicines Agency (EMA) has implemented restrictions on the use of certain quinolones in veterinary medicine, and the European Union’s updated Pharmaceutical Strategy includes provisions for environmental risk assessments of antibiotics. In the United States, the Environmental Protection Agency (EPA) and the Food and Drug Administration (FDA) are collaborating to assess the environmental impact of antibiotic residues and to update regulatory frameworks accordingly.
Looking ahead, the next few years are likely to see increased harmonization of standards and reporting requirements, as well as the integration of environmental surveillance into national AMR action plans. The One Health approach—recognizing the interconnectedness of human, animal, and environmental health—will continue to underpin policy development. International organizations are expected to provide technical support and capacity-building to help countries implement effective environmental monitoring and stewardship programs, aiming to curb the spread of quinolone resistance in environmental bacteria.
Emerging Technologies and Future Solutions
The ongoing challenge of quinolone resistance in environmental bacteria has prompted a surge in research and development of innovative technologies and strategies aimed at curbing the spread and impact of resistance genes. As of 2025, several emerging solutions are being explored, with a focus on both detection and mitigation.
One of the most promising technological advances is the deployment of next-generation sequencing (NGS) platforms for environmental surveillance. These high-throughput systems enable rapid identification of quinolone resistance genes (qnr, aac(6’)-Ib-cr, qepA, etc.) in complex environmental samples, such as wastewater, agricultural runoff, and surface waters. The integration of NGS with advanced bioinformatics pipelines allows for real-time monitoring of resistance gene dissemination, supporting early intervention strategies. Organizations such as the Centers for Disease Control and Prevention and the World Health Organization are actively promoting the adoption of genomic surveillance frameworks to track antimicrobial resistance (AMR) globally.
Another area of innovation is the development of novel water treatment technologies designed to degrade residual quinolones and reduce the selective pressure that drives resistance. Advanced oxidation processes (AOPs), including photocatalysis and ozonation, are being piloted in municipal and industrial wastewater treatment plants. These methods have demonstrated efficacy in breaking down persistent quinolone compounds, thereby limiting their environmental impact and the subsequent selection for resistant bacteria. The United States Environmental Protection Agency is supporting research into scalable AOPs and their integration into existing treatment infrastructure.
Bioremediation approaches are also gaining traction, with engineered microbial consortia and enzymes being investigated for their ability to degrade quinolones in situ. Synthetic biology tools are enabling the design of bacteria capable of metabolizing quinolones without acquiring resistance genes, offering a targeted and sustainable remediation strategy.
Looking ahead, the convergence of digital technologies, such as artificial intelligence (AI) and machine learning, with environmental microbiology is expected to enhance predictive modeling of resistance emergence and spread. These tools can analyze large datasets from environmental monitoring, antibiotic usage, and resistance gene prevalence to inform risk assessments and guide policy interventions.
International collaboration remains crucial. Initiatives like the Global Antimicrobial Resistance Surveillance System (GLASS) by the World Health Organization are expanding their scope to include environmental reservoirs, fostering data sharing and harmonized methodologies. Over the next few years, the integration of these emerging technologies and collaborative frameworks is anticipated to significantly strengthen the global response to quinolone resistance in environmental bacteria.
Forecasting the Future: Trends, Public Awareness, and the Road Ahead
As we move into 2025, quinolone resistance in environmental bacteria is recognized as a critical and growing threat to global public health. Quinolones, a class of broad-spectrum antibiotics, have been widely used in human medicine, veterinary practice, and agriculture. Their extensive application has led to the proliferation of quinolone-resistant bacteria in diverse environmental reservoirs, including water bodies, soil, and wildlife. Recent surveillance data indicate that resistance genes, such as qnr, aac(6’)-Ib-cr, and mutations in gyrA and parC, are increasingly detected in environmental isolates, often at levels paralleling or exceeding those found in clinical settings.
In 2025, several international organizations, including the World Health Organization (WHO) and the Food and Agriculture Organization of the United Nations (FAO), have intensified their monitoring and reporting efforts. These bodies emphasize the interconnectedness of environmental, animal, and human health—a concept central to the One Health approach. The WHO’s Global Antimicrobial Resistance Surveillance System (GLASS) has expanded its environmental monitoring modules, providing more granular data on the prevalence and spread of quinolone resistance genes in aquatic and terrestrial environments.
Recent studies highlight that wastewater treatment plants, agricultural runoff, and pharmaceutical manufacturing effluents remain significant sources of quinolone-resistant bacteria and resistance genes. In 2025, regulatory agencies in several countries are piloting stricter effluent standards and promoting advanced treatment technologies, such as ozonation and membrane filtration, to mitigate the release of resistant bacteria into the environment. The United States Environmental Protection Agency (EPA) and the European Medicines Agency (EMA) are among the authorities updating guidelines for environmental risk assessments of antibiotics.
Public awareness campaigns are also gaining momentum. Educational initiatives led by the Centers for Disease Control and Prevention (CDC) and WHO aim to inform the public and stakeholders about the environmental dimensions of antimicrobial resistance (AMR), including the risks associated with improper disposal of antibiotics and the importance of responsible use in agriculture and healthcare.
Looking ahead, the next few years are expected to see increased investment in environmental surveillance, the development of rapid detection technologies for resistance genes, and the implementation of integrated AMR action plans. However, challenges remain, including the need for harmonized global standards, improved data sharing, and sustained political and financial commitment. The trajectory of quinolone resistance in environmental bacteria will depend on the effectiveness of these coordinated efforts and the ability to translate scientific knowledge into actionable policy.
Sources & References
- World Health Organization
- European Medicines Agency
- Centers for Disease Control and Prevention
- European Food Safety Authority
- Food and Agriculture Organization of the United Nations
- World Health Organization
- Centers for Disease Control and Prevention
- European Centre for Disease Prevention and Control